In analytical chemistry, quantification of small molecules (metabolites, hormones, toxins) often involves mass spectrometry and isotopic dilution. Over the last 10 years, this analytical principal has been progressively adapted to the study of proteins. Today, the latest developments in the field of proteomics analysis make it possible to determine protein concentrations in biological samples using this type of approach.
In 2007, the Exploring the Dynamics of Proteomes team developed the PSAQ™ method (Protein Standard Absolute Quantification) for absolute protein quantification. This method uses whole isotopically labelled proteins as quantification standards. These standards, produced in-house, can be added to biological samples at the start of the analytical procedure. They allow very precise and accurate determination of target protein concentrations. PSAQs have been used to analyze biomarkers of disease. In this context, the PSAQ
™ is a labelled analogue of the target biomarker
[1].
A new study
[2] was developed to demonstrate the reliability and advantages to be gained by combining the PSAQ™ method with mass spectrometry for the simultaneous (multiplex) assay of several protein biomarkers present in the blood.
Biomarkers of myocardial infarction (myoglobin, creatine kinases M and B, lactate dehydrogenase B and troponin I) were chosen as target proteins. Each of these biomarkers is used clinically in the diagnosis of myocardial infarction, for which they are currently assayed individually in medical analysis laboratories using immunological or enzymatic tests. We developed a procedure capable of assaying these biomarkers simultaneously. Because serum proteins are 60% albumin, 35% globulins, we first had to develop methods to pre-fractionate the serum sample. Without this step, proteins of lower abundance, among which biomarkers are found, would be masked. We then used a specific mass spectrometry method, SRM, which allows analysis to be focused on "signature peptides" (specific fragments of biomarkers). This enhances specificity and sensitivity as these signature peptides can be much more easily detected by mass spectrometry than whole proteins by any method.
A collaboration was established with cardiologists at Henri Mondor Hospital, Créteil so as to be able to test the method on clinical samples. Four biomarkers were quantified simultaneously in 14 µL serum samples from patients having suffered myocardial infarction. Troponin I, which is a very low abundance biomarker, could also be quantified, but only with 1 mL samples. Correlation between the results of this analysis and data from the medical biology laboratory was very good
[2].
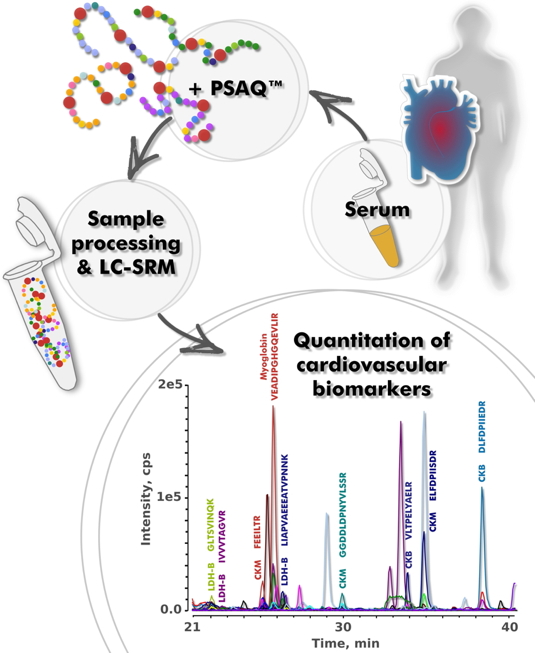
The PSAQ™-SRM technique applied for the quantification of cardiovascular biomarkers.
As a follow-up to this study, the team is currently working on developing a multiplexed analysis method to study other clinically relevant panels of biomarkers. In particular, the BIOPANEL project aims to assess the relevance of a group of biomarker candidates for diagnosis and follow-up of acute alcohol-induced hepatitis. This project was funded in 2011 by GRAVIT, and is a collaboration with the hepato-gastroenterology department at Grenoble University teaching Hospital.