 Coordinateur du projet
Coordinateur du projet :
Virginie BRUN
Responsable EDyP :
Virginie BRUN
Financement :
GRAVIT, ANR Emergence,
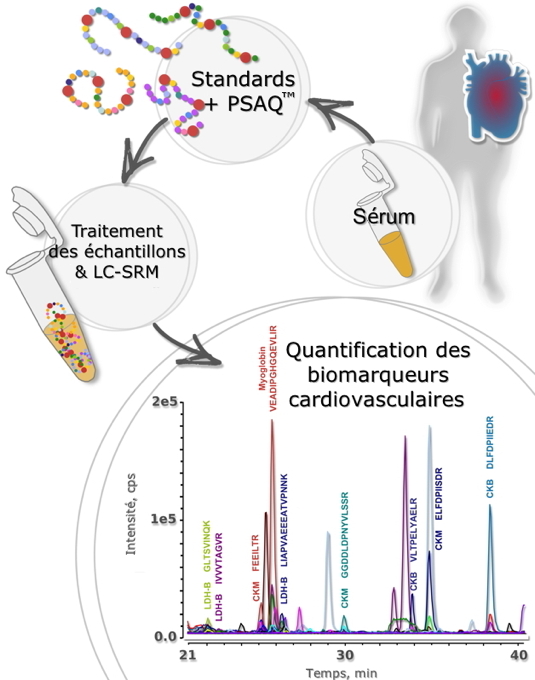
PRIME-XSPrincipe de la technique PSAQ™-SRM pour la quantification de biomarqueurs cardiovasculaires. En chimie analytique, la quantification des petites molécules (métabolites, hormones, toxiques) est souvent effectuée par spectrométrie de masse et dilution isotopique. Depuis une dizaine d’années, ce principe d’analyse a été progressivement adapté à l’étude des protéines. Aujourd’hui, les derniers développements qui ont vu le jour dans le domaine des analyses protéomiques montrent que l’on peut déterminer les concentrations des protéines dans les échantillons biologiques grâce à ce type d’approches.
En 2007, notre équipe a développé la méthode PSAQ™ (
Protein Standard Absolute Quantification) pour la quantification absolue des protéines (Voir
lettre scientifique de l’iRTSV n°11 :
PSAQ™, un mètre-étalon pour la protéomique). Cette méthode utilise des protéines entières, marquées avec des isotopes stables, comme standards de quantification. Ces standards, produits par l'équipe, sont ajoutés aux échantillons biologiques dès le début de la procédure analytique. Ils permettent de déterminer de manière très exacte et précise les concentrations des protéines cibles, par exemple des biomarqueurs de pathologies dont ils constituent des analogues marqués
[1].
L’objectif de mener une nouvelle étude
[2] était de démontrer la fiabilité et l’intérêt du couplage de la méthode PSAQ™ et de la spectrométrie de masse pour le dosage simultané (multiplexé) de plusieurs biomarqueurs protéiques présents dans le sang.
Ainsi des biomarqueurs de l’infarctus du myocarde (myoglobine, créatine kinases M et B, lactate déhydrogénase B et troponine I) ont ainsi été choisis comme protéines cibles à doser. Classiquement, chacun de ces biomarqueurs est dosé dans des laboratoires d’analyses médicales à l’aide de tests immunologiques ou enzymatiques. Nous avons développé une procédure d’analyse adaptée au dosage simultané de ces biomarqueurs. Pour ce faire, une méthode de pré-fractionnement de l’échantillon sérique a tout d’abord été mise au point afin d’éliminer les protéines les plus abondantes du sérum. Une telle étape est indispensable ; en effet, le sérum est particulièrement difficile à analyser car il contient 60% d’albumine et 35% de globulines qui masquent les protéines de plus faible abondance parmi lesquelles se trouvent les biomarqueurs. Ensuite, afin d’obtenir une détection spécifique et sensible, nous avons utilisé une méthode particulière de spectrométrie de masse, la SRM (
Selected Reaction Monitoring), qui permet de focaliser l’analyse sur des « peptides signatures » (fragments spécifiques des biomarqueurs) qui peuvent être détectés en spectrométrie de masse beaucoup plus facilement que les protéines intactes.
Afin d'évaluer cette méthode sur des échantillons cliniques, une collaboration a été établie avec des cardiologues de l’hôpital Henri Mondor de Créteil. Ainsi, quatre biomarqueurs ont pu être quantifiés simultanément chez des patients atteints d’infarctus du myocarde à partir d'échantillons de seulement 14 µl de sérum. La troponine I, un biomarqueur de très faible abondance, a pu être également quantifiée, mais en partant d’échantillons de 1 ml de sérum. Une très bonne corrélation a été obtenue entre ces résultats et les données qui étaient connues du laboratoire de biologie médicale
[2].
Suite à cette étude, l'équipe travaille actuellement au développement d’une méthode d’analyse multiplexée permettant l’étude de nouveaux panels de biomarqueurs d’intérêt clinique. En particulier, le projet BIOPANEL vise à évaluer la pertinence d’un ensemble de candidats biomarqueurs en vue du diagnostic et du suivi thérapeutique des hépatites aiguës alcooliques. Ce projet financé en 2011 par GRAVIT, est effectué en collaboration avec le service d’hépato-gastro-entérologie du CHU de Grenoble.Publications :
[1] Brun V, Dupuis A, Adrait A, Marcellin M, Thomas D, Court M, Vandenesch F and Garin J Isotope-labeled protein standards: Toward absolute quantitative proteomics.
Molecular and Cellular Proteomics, 2007
[2] Huillet C, Adrait A, Lebert D, Picard G, Trauchessec M, Louwagie M, Dupuis A, Hittinger L, Ghaleh B, Le Corvoisier P, Jaquinod M, Garin J, Bruley C and Brun V
Accurate quantification of cardiovascular biomarkers in serum using protein standard absolute quantification (PSAQ™) and selected reaction monitoring.
Molecular and Cellular Proteomics, 2012