 Technological and scientific research themes
Technological and scientific research themes Advances in molecular and cellular profiling technologies for biological materials (tissues, cell cultures) transformed Omics sciences by bringing them into the Big Data fied while taking into account the personal and sensitive aspects of these data. Within the MODAL group, we develop
bioinformatics and AI approaches to be able to start from raw omics data and go all the way to the development of predictive models in precision medicine. We apply our methodologies in research projects carried out within national and European consortia.
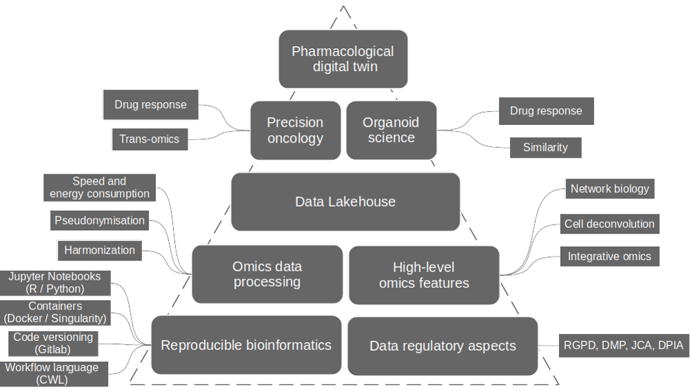 Data regulatory aspects
Data regulatory aspects - In collaboration with lawyers specialized in digital law, we ensure that the regulatory and legal aspects relating to the life cycle of the data collected during a research project comply with GDPR regulations.
Reproducible bioinformatics - We use FAIR technologies and best practices that facilitate the monitoring, reproducibility and sharing of our computer codes.
Omics data processing - We process and harmonize raw omics data taking into consideration aspects of data pseudonimization and optimization of calculations to reduce their energy footprint.
High-level omics features - To deal with the high dimensionality of omics data complicating their analysis, we use cellular deconvolution, network biology and multi-omics methodologies, relying on a priori biological knowledge, to generate higher-level features facilitating biological interpretation of the results.
Data Lakehouse - The volume, diversity and complexity of omics data requires adapting the means used to analyze and model them. We develop Data Lakehouse infrastructures dedicated to the objectives of research projects to reproduce data processing, ensure versioning, finely configure authorized access, facilitate collaborative work and allow the deployment of analytics and machine learning methods.
Precision oncology – Our
first application field rely on the accumulation of omic profiling data, from tumor tissues of patients included in clinical drug trials, to develop AI models in oncology to predict patient response to therapies. These models use the cellular and molecular characteristics of tumor tissues, integrated with the patient's clinical and biological data, to predict the clinical benefit associated with each treatment.
Organoid science – Our
second application field consists of analyzing the cellular and molecular complexity of organoids and tumor spheroids, measured from omic profiling data, in order to assess the ability of these 3D cell cultures to recapitulate the biological processes present in the corresponding reference tissues. We also work on the value of omics data generated from 3D cell cultures to enrich AI models of predictions of patient response to drugs.
Pharmacological digital twin - Within a translational research consortium, we integrate our methodologies to support the development of digital twins in pharmacology capable of helping clinicians to improve the therapeutic management of patients.
 Research projects
Research projects 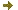 AI models predicting patient response to cancer treatment
AI models predicting patient response to cancer treatmentDrug treatments for advanced cancers have evolved considerably over the past ten years with the availability of an increasing number of targeted therapies based on protein kinase inhibitors and immunotherapies. At the same time, omics profiling technologies have made it possible to generate multi-omic data from large cohorts of patient tumors. One of the main challenges in precision medicine is now to optimize the choice of treatments for patients by combining clinical, histological and genomic information about their tumors.
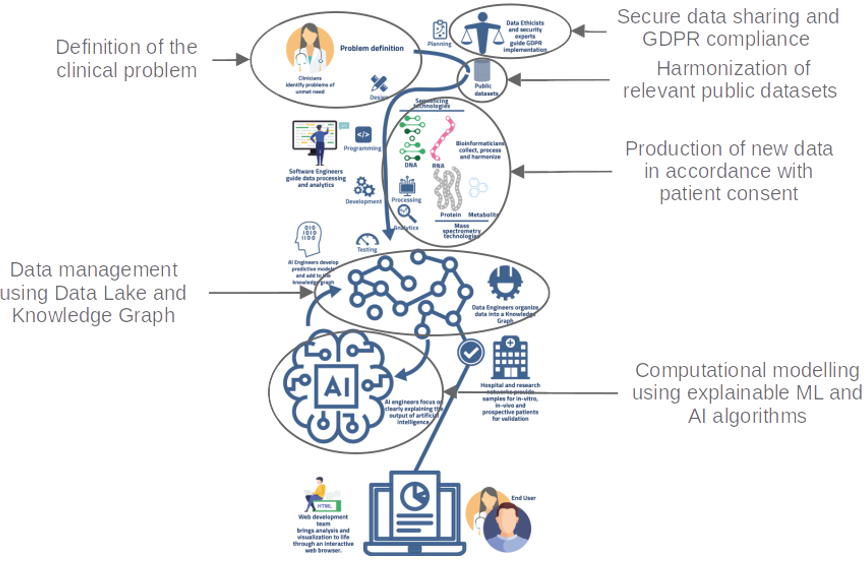
In the context of multi-disciplinary consortia, we provide our expertise at different levels of the data life cycle. First, we homogeneously reprocess omics data and generate high-level biological features to enrich this data. Then, we facilitate collaborative work and governance of data collections by organizing them within Data Lake infrastructure. Finally, we develop methodologies integrating bioinformatics and machine learning algorithms capable of predicting the response to anti-tumor treatments of patients with cancer and identifying the cellular and molecular signatures supporting these predictions.
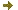 Modeling of cellular and molecular similarity between organoids / tumor spheroids and tissues
Modeling of cellular and molecular similarity between organoids / tumor spheroids and tissues - 3D cell cultures, such as organoids and tumor spheroids, have become biological tools arousing great interest because they allow a better understanding of the complexity of tissues, in addition to 2D cell cultures. This has led to their use to study human pathologies, through gene editing, and for screening therapeutic molecules.
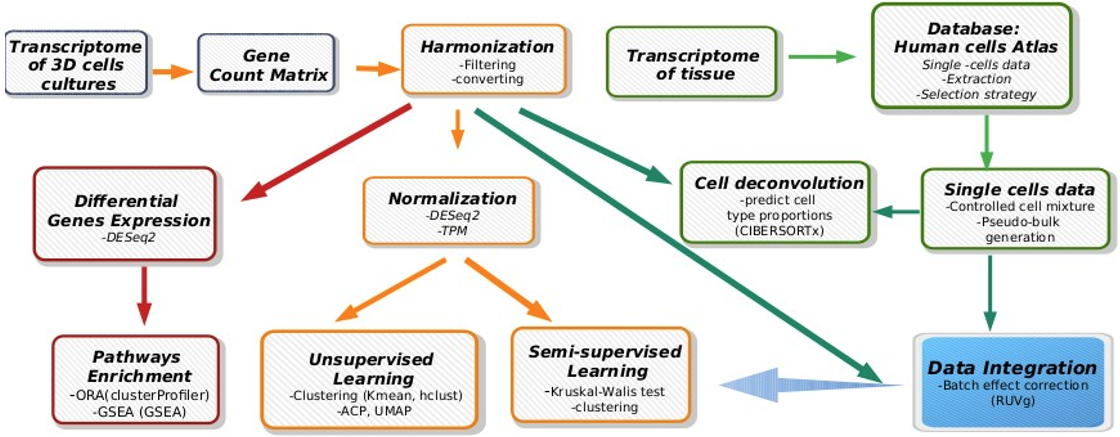
In collaboration with laboratories improving 3D cell cultures, we support the maturation of these new biological avatars by providing bioinformatics and machine learning methodologies to finely study the degree of fidelity between the desired tissue phenotype and 3D cell cultures, to better understand the mechanisms of action of therapeutic molecules and to assess the value of these new cellular models in predicting patient response to treatments.
 Group members
Group members
 Alumni
Alumni
- Damien Alouges (AI engineer)
- Thierno Sidy Bah (M2 student by apprenticeship)
- Elodine Coquelet (Engineer student)
- Pauline Bazelle (Bioinformatics engineer)
- Solène Mauger (M2 student)
- Etienne Bardet (M2 student)
- Evan Seffar (M2 student)
- Islam El Boudi (M1 student)
- Bruno Giotti (Postdoctoral researcher)
- Georges Bedran (M2 student)