The continuing development of methods and technologies for the large-scale characterisation of biological samples is a double-edged sword for researchers in molecular biology: on the one hand, these provide reliable and rapid access to thousands of genes, transcripts, proteins or metabolites, making it possible to test a considerable number of hypotheses concerning the functioning of living organisms. On the other hand, the multiplication of hypotheses that can be studied simultaneously increases the risk that one of them is incorrectly validated by chance: a so-called false discovery.
This increase in the risk of false discoveries roots in combinatorics: the probability is low that a randomly chosen biomolecule display measurement fluctuations that match the expectations induced by the hypothesis being studied. However, if several thousand biomolecules are considered simultaneously, the probability that at least one of them behaves accordingly becomes significant.
Controlling the risk for false discoveries is therefore a major challenge for modern biology, and advanced statistical methods are needed. However, the gap between the theoretical framework for handling this statistical risk and the complexity of the experimental designs actually used in biology is so wide that it can compromise the ability to effectively control for the risk of false discoveries. This gap is particularly important in
proteomics, because of the complexity of the measurement achieved by coupling mass spectrometry and liquid chromatography, and because of the small number of samples that can generally be analysed for a given experiment.
To overcome this, researchers at BGE/EDYP have been working for a number of years on putting into perspective the experimental constraints and theoretical hypotheses needed to control for false discoveries, so that they can be integrated into the data analysis workflows [4] and implemented into different software suites (e.g. www.prostar-proteomics.org ) to refine quality control tools. Their recent work has focused on the theory of Knockoff filters, which has recently revolutionised the field of
selective inference, by proposing to use random draws to better characterise the properties of false discoveries. In particular, this team has made the link between the use of Knockoffs and the empirical methods used in proteomics [2] to propose innovative methods [3,
1].
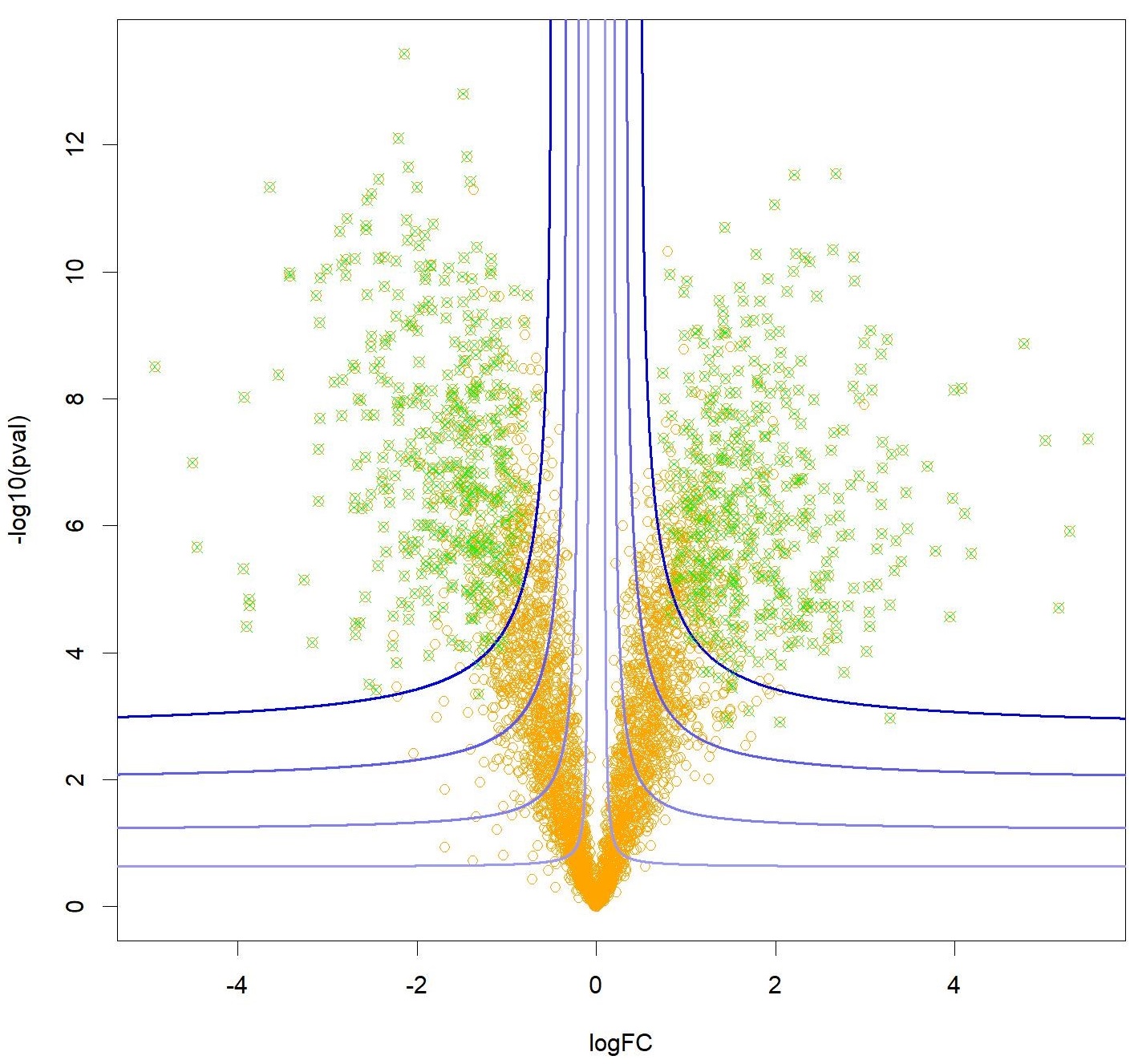
Figure: A volcano plot with orange dots representing the proteins that can explain a difference in phenotype according to their significance (on the Y-axis) and the size of the effect measured (X-axis). Knockoff filters are used to control for the false discovery rate associated with a selection of proteins (in green) following a hyperbolic decision frontier (in blue), allowing for both the effect size and the significance to be accounted for.
Proteomics: Large-scale characterisation (identification and quantification) of proteins present in a biological sample.
Selective inference: A field of high-dimensional statistics which deals with the generalisation of knowledge drawn from experimental data where the data have been previously selected for their specific characteristics.
Funds
- Multidisciplinary Institute in Artificial Intelligence (MIAI @ Grenoble Alpes, ANR)
- Programme GRAL via Chemistry Biology Health Graduate School at University Grenoble Alpes (ANR)
- ProFI (Proteomics French Infrastructure, ANR